Software
- OTRIMLE: robust model-based clustering
- RSC: robust and sparse correlation matrix
- hyQSAR: hybrid quantitative structure–activity relationship (QSAR) modelling
- RCPS: robust clustering for patient subtyping
OTRIMLE: robust model-based clustering
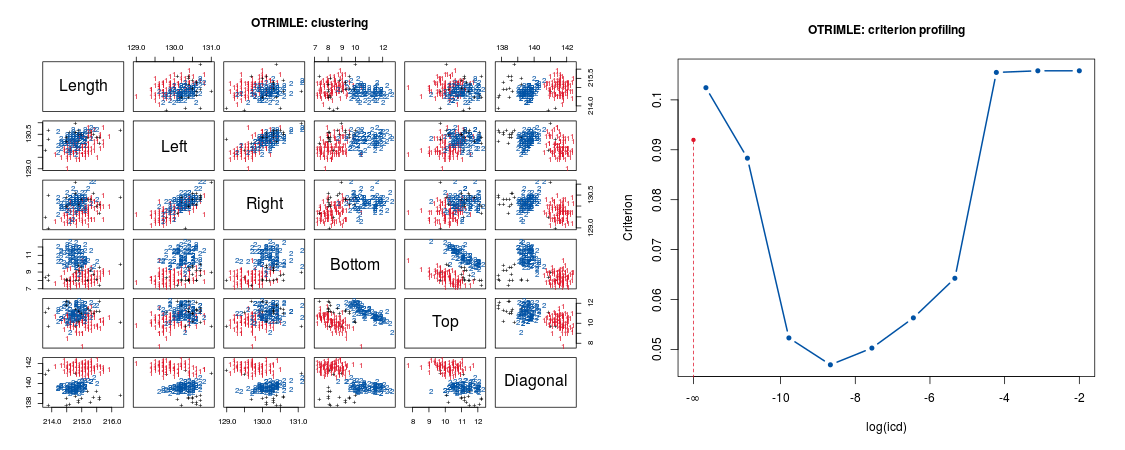
OTRIMLE is an R package that performs robust cluster analysis allowing for outliers and noise that cannot be fitted by any cluster. The data are modelled by a mixture of Gaussian distributions and a noise component, which is an improper uniform distribution covering the whole Euclidean space. Parameters are estimated by (pseudo) maximum likelihood. This is fitted by a EM-type algorithm.
References
- P. Coretto and C. Hennig (2016). Robust improper maximum likelihood: tuning, computation, and a comparison with other methods for robust Gaussian clustering. Journal of the American Statistical Association, Vol. 111(516), pp. 1648-1659. DOI link (open access)
- P. Coretto and C. Hennig (2107). Consistency, breakdown robustness, and algorithms for robust improper maximum likelihood clustering. Journal of Machine Learning Research, Vol. 18(142), pp. 1-39. Download (open access)
RSC: robust and sparse correlation matrix
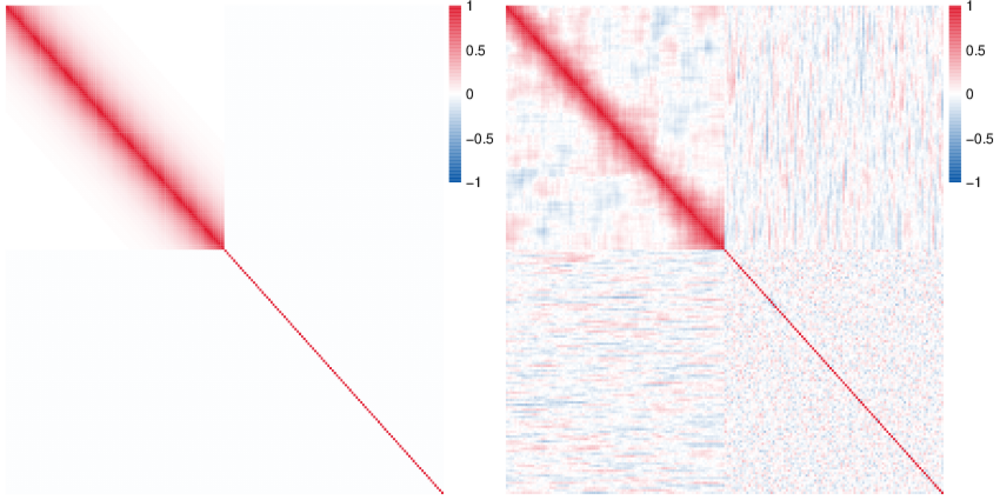
RSC is an R package that computes an estimate of the correlation matrix that is resistant to large amount of noise/outliers. Adaptive thresholding is applied to obtain a sparse estimate. The software is fairly scalable and can handle massive data sets.
References
A. Serra, P. Coretto, M. Fratello, and R. Tagliaferri (2018). Robust and sparse correlation matrix estimation for the analysis of high-dimensional genomics data. Bioinformatics, Vol. 34(4), pp. 625-634. DOI link.
hyQSAR: hybrid quantitative structure–activity relationship (QSAR) modelling
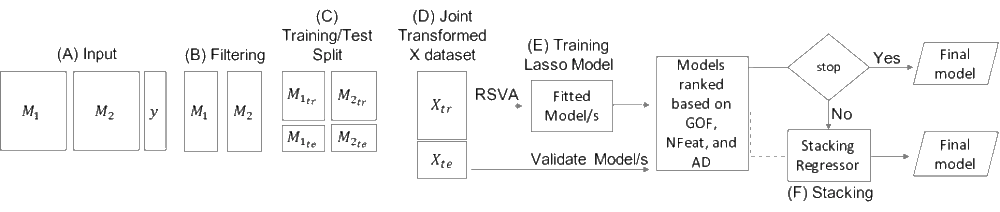
hyQSAR is an R package that can be used to perform hybrid QSAR modelling by integrating structural properties of chemical compounds and their molecular mechanism-of-action (MOA) information. HyQSAR enables easier interpretation of the mechanistic associations between the exposure properties and their biological effects for a given endpoint of interest. It implements a LASSO-based method to optimise the feature selection from multiple data sources as well as the best model parameters by random split validation. In addition, hyQSAR allows automatic model validation according to the OECD requirements. Multiple visualisations are also provided to help the interpretation of the output.
Download software (development version) via GitHub (also contains data used in the references given below)
References
A. Serra, S. Önlü, P. Coretto, and D. Greco (2019). An integrated quantitative structure and mechanism of action-activity relationship model of human serum albumin binding. Journal of Cheminformatics, Vol. 11(38), pp.1-10. DOI link (open access)
RCPS: robust clustering for patient subtyping
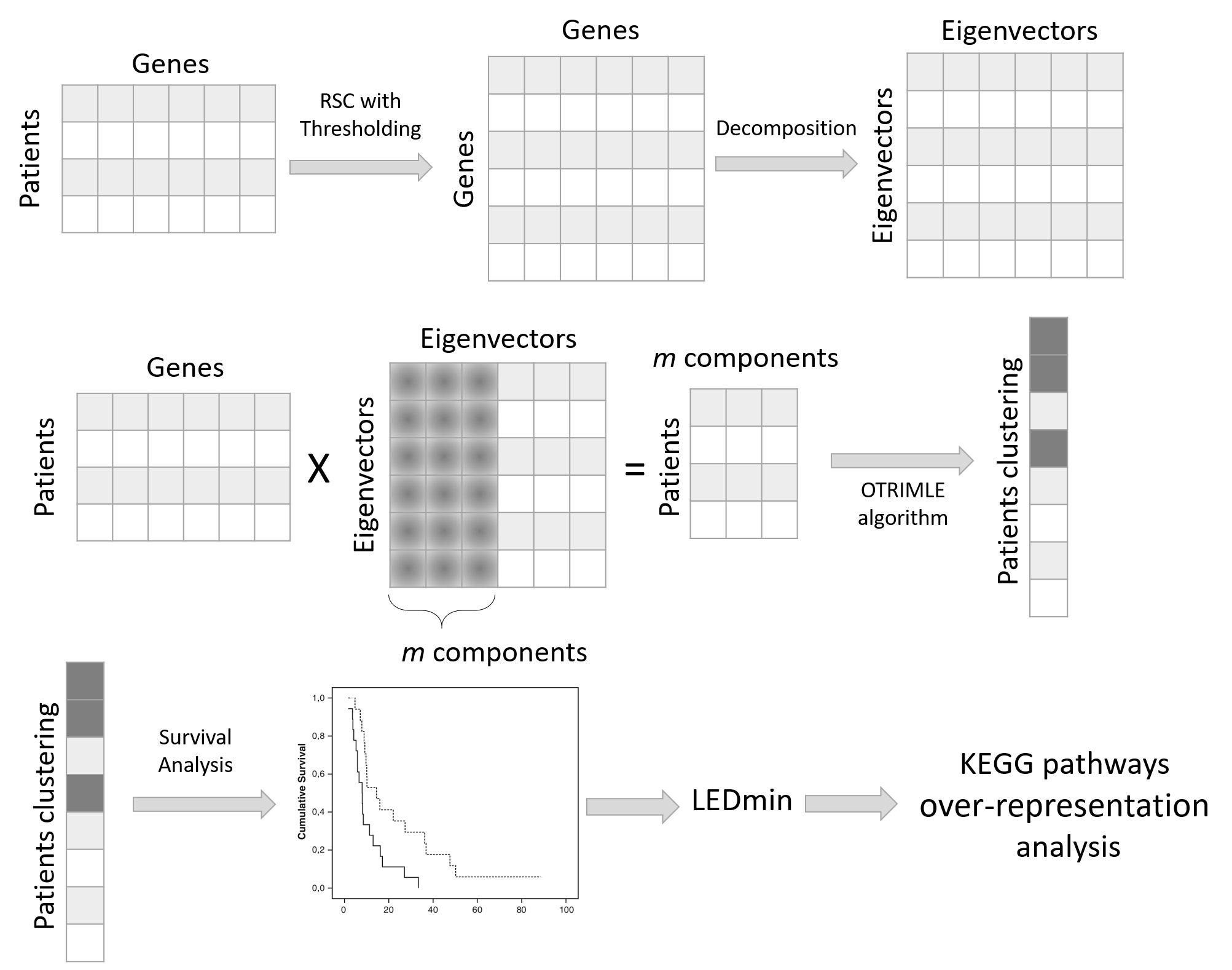
RCPS is an R library that performs robust clustering for patient subtyping that guarantees optimal separation in terms of survival patterns. The method computes a robust and sparse correlation matrix of the genes, then decomposes it and projects the patient data onto the first m spectral components of this matrix. After that, the OTRIMLE, a robust and adaptive to noise clustering algorithm is applied. The clustering is set up to optimize the separation between survival curves estimated cluster-wise.
Download software (development version) via GitHub (also contains data used in the references given below)
References
P. Coretto, A. Serra and R. Tagliaferri (2018). Robust clustering of noisy high-dimensional gene expression data for patients subtyping. Bioinformatics, Vol. 34(23), pp. 4064–4072. DOI link
| home |